Section: New Results
Computational Anatomy
Statistical Analysis of Diffusion Tensor Images of the Brain
Participants : Marco Lorenzi [Correspondent] , Nicholas Ayache, Xavier Pennec.
Image non-linear registration, Longitudinal modeling, Alzheimer's disease
Alzheimer's disease is characterized by the co-occurrence of different phenomena, starting from the deposition of amyloid plaques and neurofibrillary tangles, to the progressive synaptic, neuronal and axonal damages. The brain atrophy is a sensitive marker of disease progression from pre-clinical to the pathological stages, and computational methods for the analysis of magnetic resonance images of the brain are currently used for group-wise (cross-sectional) and longitudinal studies of pathological morphological changes in clinical populations. The aim of this project is to develop robust and effective computational instruments for the analysis of longitudinal brain changes. In particular novel methods based on non-linear diffeomorphic registration have been investigated in order to reliably detect and statistically analyze pathological morphological changes [5] (see Fig.8 ). This project is also focused in the comparison of the trajectories of longitudinal morphological changes [31] estimated in different patients. This is a central topic for the development of statistical atlases of the longitudinal evolution of brain atrophy.
|
Statistical Learning via Synthesis of Medical Images
Participants : Hervé Lombaert [Correspondent] , Nicholas Ayache, Antonio Criminisi.
This work has been partly supported by a grant from Microsoft Research-Inria Joint Centre, by ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images)
Statistical learning, Synthesis
Machine learning approaches typically require large training datasets in order to capture as much variability as possible. Application of conventional learning methods on medical images is difficult due to the large variability that exists among patients, pathologies, and image acquisitions. The project aims at exploring how realistic image synthesis could be used, and improve existing machine learning methods.
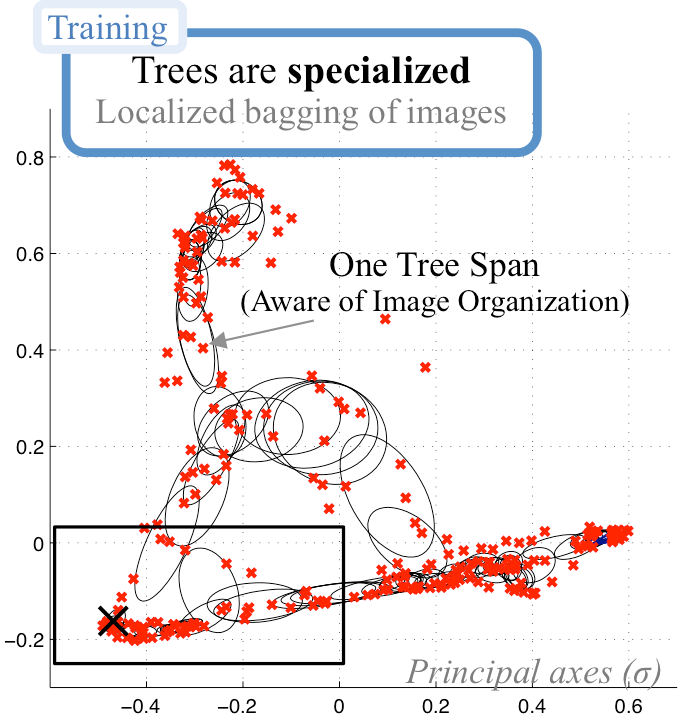
First year tackled the problem of better exploiting existing training sets, via a smart modeling of the image space (Fig. 9 ), and applying conventional random forests using guided bagging [21] . Synthesis of complex data, such as cardiac diffusion images (DTI), was also done. Synthesis of complex shapes, using spectral graph decompositions, is currently on-going work.
The modeling of shapes also includes novel representations based on the spectral decomposition of images[4] which are more robust to large deformations when comparing multiple patients.
|
Statistical analysis of heart shape, deformation and motion
Participants : Marc-Michel Rohé [correspondent] , Xavier Pennec, Maxime Sermesant.
This work was partly supported by the FP7 European project MD-Paedigree and by ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images)
Statistical analysis, Registration, Reduced order models, Machine learning
The work aims at developping statistical tools to analyse cardiac shape, deformation, and motion. In particular, we are interested in developping reduced order models so that the variability within a population described by a complex model can be reduced into few parameters or modes that are clinically relevant. We use these modes to represent the variability seen in a population and to relate this variability with clinical parameters, and we build group-wise statistics which relate these modes to a given pathology. We focus on cardiomyopathies and the cardiovascular disease risk in obese children and adolescents.
Geometric statistics for Computational Anatomy
Participants : Nina Miolane [Correspondent] , Xavier Pennec.
Lie groups, pseudo-Riemannian, Statistics, Computational Anatomy
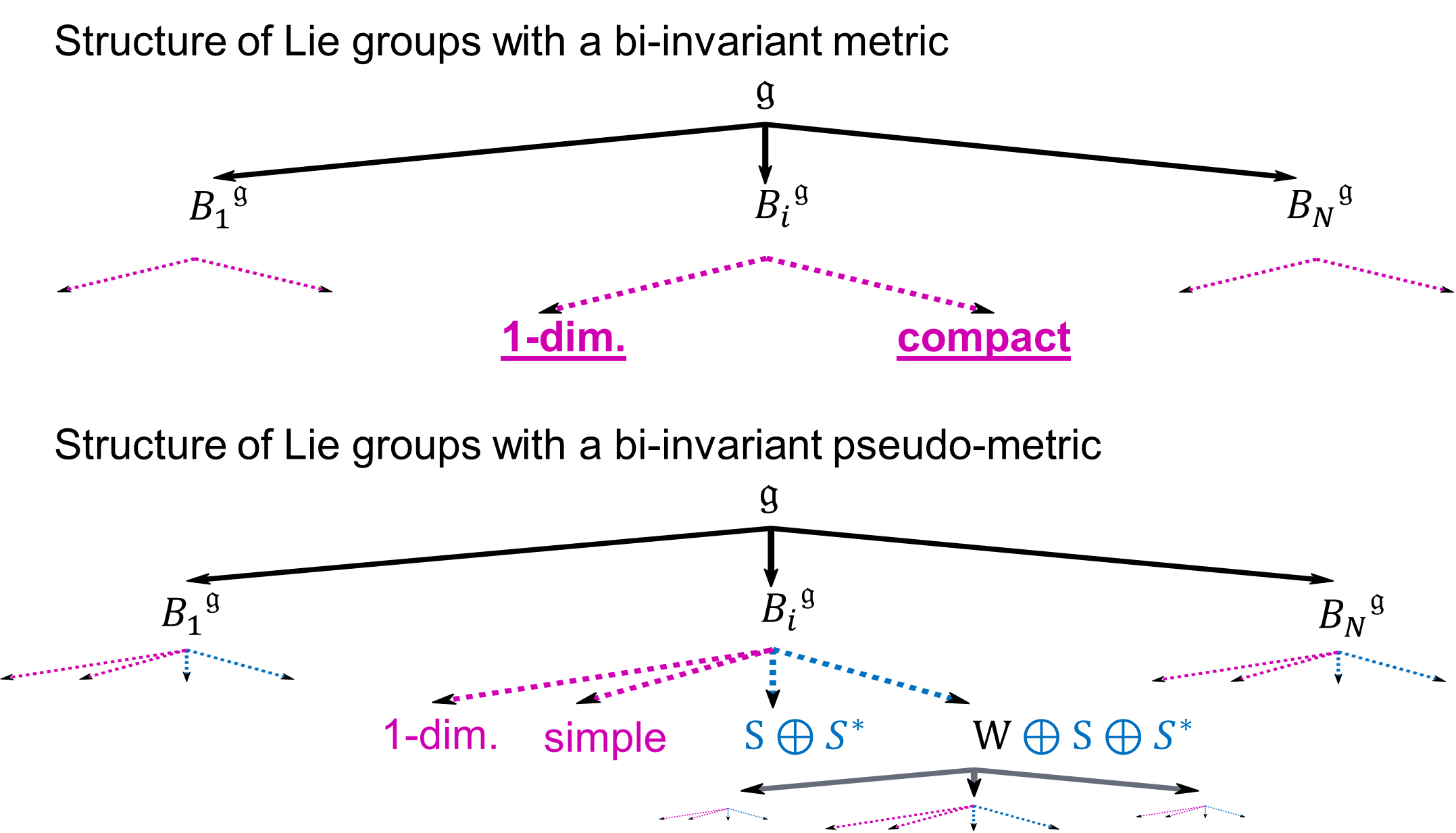
Lie groups are widely used in mathematical models for Medical Imaging. In Computational Anatomy for example, an organ's shape can be modeled as the deformation of a reference shape, in other words : as an element of a Lie group. If one wants to analyze the variability of the human anatomy, e.g. to help diagnose diseases, one has to perform statistics on Lie groups. We investigate the geometric structures on Lie groups that enable to define consistent statistics. A Lie group is a manifold with an additional group structure. Statistics on Riemannian manifolds have be studied throughout the past years. One may wonder if we could use the theory of statistics on Riemannian manifolds for statistics on . To this aim, we need to define a Riemannian metric on the Lie group that is compatible with the group structure: a so-called bi-invariant metric. However, it is known that most Lie groups do not admit any bi-invariant metric. One may wonder if we could generalize the theory of statistics on Riemannian manifolds to pseudo-Riemannian manifolds and use it for statistics on . To this aim, we need to define a bi-invariant pseudo-metric on . How many Lie groups do admit such a pseudo-metric and can we compute it? These investigations and their results (see Fig. 10 ) were presented at MaxEnt 2014 [24] .
|
Statistical Analysis of Diffusion Tensor Images of the Brain
Participants : Vikash Gupta [correspondent] , Nicholas Ayache, Xavier Pennec.
Population specific multimodal brain atlas for statistical analysis of white matter tracts on clinical DTI.
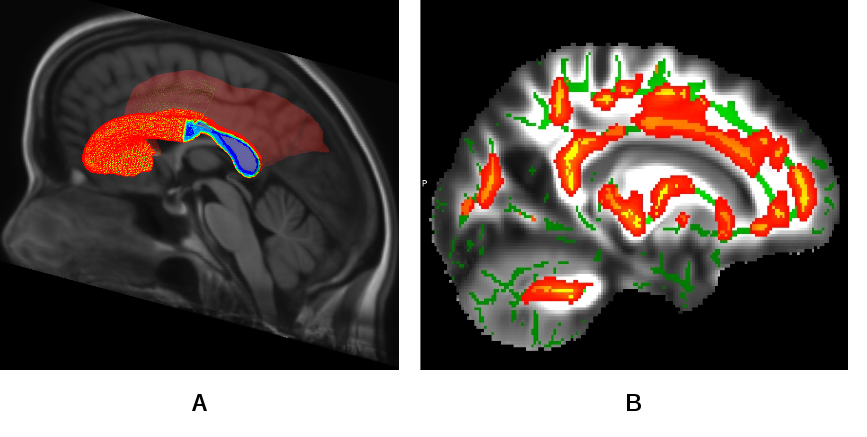
HIV virus can cross the hematoencephallic barrier and affect the neural connectivity in the human brain causing compromised motor controls, loss in episodic, long term memory and working memory, loss in attention/concentration and visual agnosia. These cognitive losses are characterized by the neuropsychological (NP) test scores and believed to be correlated with destruction of white matter (WM) integrity among the HIV patients. For quantifying the loss in WM integrity, the HIV subjects are compared against controls using a tract based spatial statistics (TBSS) routine. The standard TBSS routines uses univariate statistics using the fractional anisotropy (FA) maps. However, we improved on the existing routines using tensor based registration for normalizing the diffusion tensor images (DTI) followed by a multivariate statistics using the full tensor information. With the improved method it is possible to detect differences in WM regions which was not possible using the existing TBSS routines. For this study a population specific multimodal (T1 and DTI) brain atlas was developed from the population. The joint atlas also contains a probabilistic parcellation of WM regions in the brain which can be used for region of interest (ROI) based statistical studies (see Fig.11 ).
|
Longitudinal Analysis and Modeling of Brain Development
Participants : Mehdi Hadj Hamou [correspondent] , Xavier Pennec, Nicholas Ayache.
This work is partly funded through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain development, adolescence, longitudinal analysis, non-rigid registration algorithm, extrapolation, interpolation
This work is divided into 2 complementary studies about longitudinal trajectories modeling:
-
Diffeomorphic registration parametrized by Stationary Velocity Fields (SVF) is a promising tool already applied to model longitudinal changes in Alzheimer's disease. However, the validity of these model assumptions in faithfully describing the observed anatomical evolution needs to be further investigated. In this work, we thus analyzed the effectiveness of linear regression of SVFs in describing anatomical deformations estimated from past and future observations of the MRIs.
-
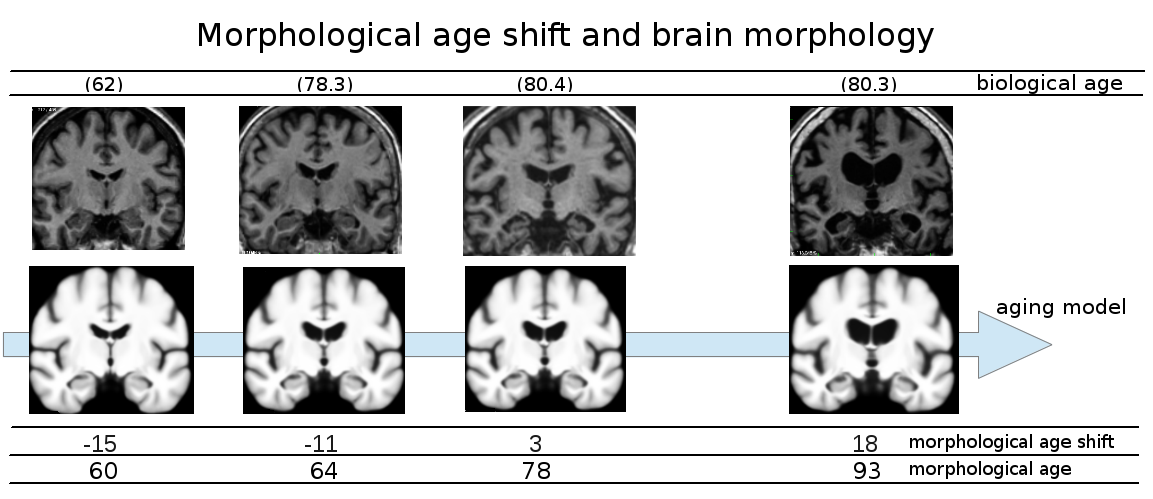
Due to the lack of tools to capture the subtle changes in the brain, little is known about its development during adolescence. The aim of this project is to provide quantification and models of brain development during adolescence based on diffeomorphic registration parametrized by SVFs (see Fig.12 ). We particularly focused our study on the link between gender and the longitudinal evolution of the brain. This work was done in collaboration with J.L. Martinot et H. Lemaître (Inserm U1000).